How to handle hospital-acquired infections with genomics
Technology used to analyse the human genome can be applied to infective agents to identify and track the outbreak
Our new-found (and still developing) capacity to determine and analyse human genome sequences has profound implications for medicine. In fact, it is not only human genomics that can improve healthcare. DNA code can now be swiftly deciphered for simpler lifeforms, too.
Indeed, it is in many ways less technically challenging, thanks to their much smaller genomes. And it is easy to see why knowing more about the genomes of fungal, bacterial and viral agents of disease (pathogens) could be important for their control and management.
Tackling HAIs
Prevention and treatment of hospital-acquired infections, or HAIs (also sometimes called nosocomial infections), is a constant battle for hospital staff, and the rising spectre of antibiotic resistance makes vigilance against outbreaks and swift action to halt them ever more important. Not only do HAIs represent a serious cause of patient morbidity and mortality, they also place significant burdens on hospitals. So can genomics be put to work to lighten the load?
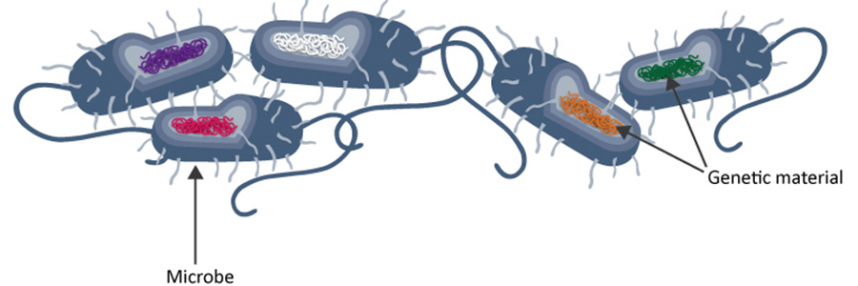
Pathogen genomes can potentially reveal important information over and above that available from traditional microbiological investigations, which can readily identify the pathogens present and determine their sensitivity – or, increasingly, resistance – to specific antibiotics.
However, different isolates of apparently identical pathogens may in fact harbour key differences at the genomic level, providing new opportunities to understand and tackle HAI outbreaks.
Insight into sources of infection
This was strikingly illustrated by a case study reported last year, using whole genome sequencing techniques to investigate infections in a new hospital burns unit. Burns patients are especially vulnerable to infections and sepsis, which are a leading cause of death in this group. Burns units therefore employ especially stringent infection control measures to minimise risks.
Researchers in the new burns unit used genome sequencing to investigate a number of Pseudomonas aeruginosa infections. P. aeruginosa is a common bacterium that rarely troubles healthy individuals, but can result in a range of infections in immunosuppressed patients, including those with burns. It can come from medical devices such as catheters, and also water sources; as burns treatment often involves hydrotherapy, it is particularly important to ensure that the water supplies are not contaminated.
The research tested both patients and regions of the burns unit regularly for P. aeruginosa; samples from infected patients and the environment were used for genomic analysis and comparison. Some of the bacteria from infected patients proved to be genetically identical to those in the hydrotherapy systems; after new and even more rigorous infection control measures for these systems were put in place, no further patient infections were detected.
Similar examples continue to emerge; for example, using a new prototype miniature genome sequencer for rapid analysis of a Salmonella outbreak at a Birmingham hospital, an expert was able to determine that there were two elements within the outbreak, one eventually traced back to contaminated food supplies and another from travel abroad.
Find out more
These are valuable demonstrations of how technologies developed to decipher the health secrets of the human genome could also be put to use for infective agents.
To learn more about the potential impacts of genomics in healthcare, including in diagnosing and tracking infections, try our introductory online course.
–