What’s in a little variation?
Genomic variation is the root of many of our traits, including what we look like and our susceptibility to many diseases. But what is it?
We are all built to the same basic human blueprint and have most of our genomic information in common. Nevertheless, variation between individuals does exist. This can be in clearly visible traits like eye colour and height, but also in less visible ways, such as our susceptibility to certain diseases.
This diversity is underpinned by differences in our genomic make-up, known as genomic variation. This variation not only determines our uniqueness but is crucial to our success as a species. It provides the raw material for natural selection, helping us to adapt to our changing environment over time as we have evolved.
So, just how similar are we?
On average if the genomes of two humans were compared, they would be 99.8– 99.9% the same. This sounds very similar, but the human genome is very big so the 0.1– 0.2% that differs includes roughly three to five million differences. These differences are called variants.
Some genomic variants affect susceptibility to particular diseases, others directly result in disease, while many others are benign.
What sort of variants do we find when we study human genomes?
There are many different types of variant in our individual genomes. Some even exist outside of the DNA sequence itself. Below, we’ll look at some of the different types you might find.
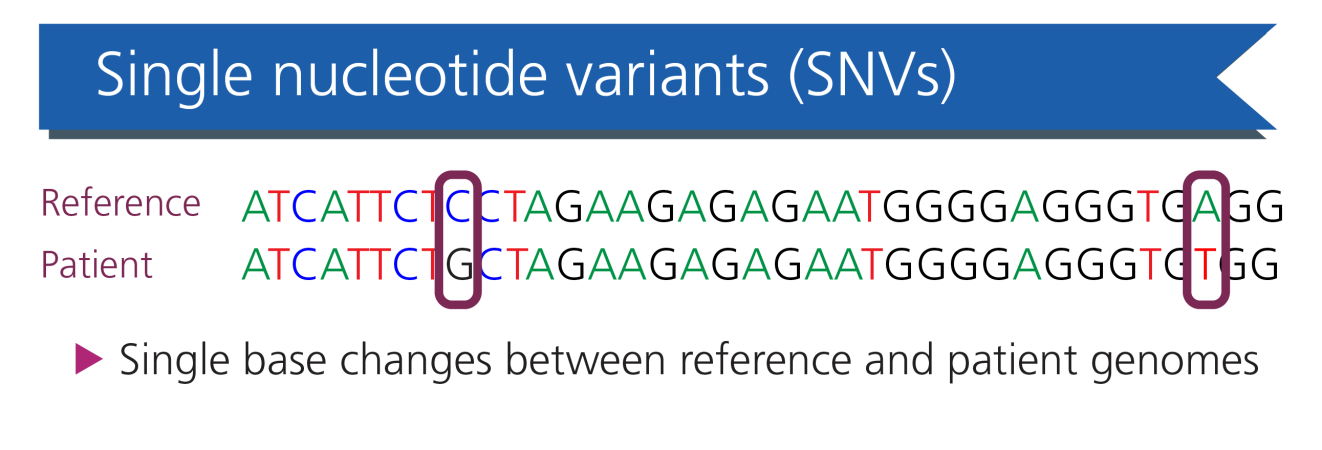
Single nucleotide variants (SNVs): the most common variant, in which one single base is substituted for another. The most common SNVs, present in the population at a frequency of greater than 1%, are further defined as single nucleotide polymorphisms.

Indels: the second most common variant, in which a short string of nucleotides become inserted and/or deleted from the DNA sequence.

Short tandem repeats (STRs): DNA sequences of between two and six nucleotides repeated alongside each other. Together, STRs comprise around 3% of our DNA. The number of repeats at a given locus is highly variable between people, and this can be exploited for certain genomic technologies, such as linkage analysis. STR regions located within genes may become pathologically expanded, causing a variety of neurodegenerative diseases called nucleotide repeat expansion disorders.
Structural variants: large duplications or deletions of genomic DNA (typically greater than one kilobase) are known as copy number variants (CNVs). CNVs, along with large insertions and genomic rearrangements such as translocations and inversions, comprise the structural variation in the genome.
Epigenetic modifications: variants that do not affect the DNA sequence itself, such as DNA methylation and chromatin remodelling. These variants can determine when certain genes are ‘switched on’ or ‘switched off’ as and when needed. Many of these modifications are harmless but some can lead to diseases, including imprinting disorders.
Remember, not all genomic variation occurs between individuals – sometimes it occurs within one person when variants arise in the developing embryo or as they age. This can lead to a subset of cells having a different genomic make-up to the rest of the body, a situation called mosaicism. In rare disease, mosaicism can lead to a different (often milder) clinical presentation and can make diagnosis more technically challenging.
But where do these variants come from?
The 1,000 Genomes Project Consortium undertook whole genome sequencing of a diverse set of individuals from a range of populations. It found that a ‘typical’ human genome contains between four and five million variants that differ from the reference genome of which around 99.9% of which are SNVs and small indels. The consortium also found that the typical human genome had, on average, 2,100 to 2,500 structural variants. It concluded that, while fewer in number, structural variants collectively affect around 20 million bases in an average human genome.
Most of these four to five million variants are inherited from our parents, but new de novo variants can arise spontaneously, and we each have between 70 and 80 of them.
As is often the case in genomics, one small change has the potential to make a big impact – both for the patient and their family, as well as the clinicians working within the care pathway. If you would like to learn more about testing for genomic variants, you may be interested in the following courses:
- Genomics 101: Investigating the Genome Part 2: The Tests (eLearning for Healthcare, 30 minutes)
- Genomics in the NHS: A Clinician’s Guide to Genomic Testing for Rare Disease (FutureLearn, two weeks)









