Circulating tumour DNA for cancer patients
Circulating tumour DNA is commonly taken as a liquid biopsy to investigate a cancer. At present, the most common liquid biopsy in clinical practice and research is circulating tumour DNA from the peripheral blood.
Overview
DNA fragments shed into the peripheral blood are known as cell-free DNA. They are released primarily from circulating haematological cells during cell necrosis and apoptosis, and increased levels are seen with infection, inflammation and strenuous exercise, as well as in malignancy. Circulating tumour DNA (ctDNA) refers to cell-free DNA fragments derived from a cancer. Fragments of ctDNA are typically shorter in length than physiological cell-free DNA, usually around 145 base pairs (bp) compared to 166bp.
Liquid biopsy is a generic term referring to analysis of tumour-derived analytes in bodily fluids, such as peripheral blood, urine, stool, saliva, pleural fluid, ascitic fluid and cerebrospinal fluid, to provide molecular information about a tumour.
The analysis of a ctDNA liquid biopsy can be used to gather information about:
- circulating cancer cells;
- epigenetic modifications;
- point mutations;
- translocations;
- amplifications and deletions;
- chromosomal irregularities;
- protein expression and phosphorylation; and
- in vivo/in vitro culture.
Benefits of ctDNA liquid biopsy
ctDNA offers a novel method of cancer investigation. Sampling is less invasive than traditional surgical biopsies, avoiding the need for high-risk procedures. This is of particular importance for more inaccessible cancer primaries, such as those of the gastrointestinal and hepatobiliary tracts. In addition, repeat liquid biopsies are more acceptable to patients than tissue biopsies, permitting longitudinal analysis of the genomic profile of cancer over time.
ctDNA has a short half-life (less than two hours) in the circulation, so it may provide real-time dynamic information about tumour status.
Tumours display intra-tumour heterogeneity, with subclones carrying varying genomic profiles. Metastases can also display differing genomic profiles owing to clonal evolution. Liquid biopsy could theoretically overcome this spatial heterogeneity, providing a comprehensive overview of the complete molecular profile of the disease. Variants of constitutional (germline) origin as well as somatic (tumour) origin may be identified by analysis of plasma. Distinguishing between variants of constitutional and somatic origin can be achieved by paired analysis of the buffy coat (see figure 1).
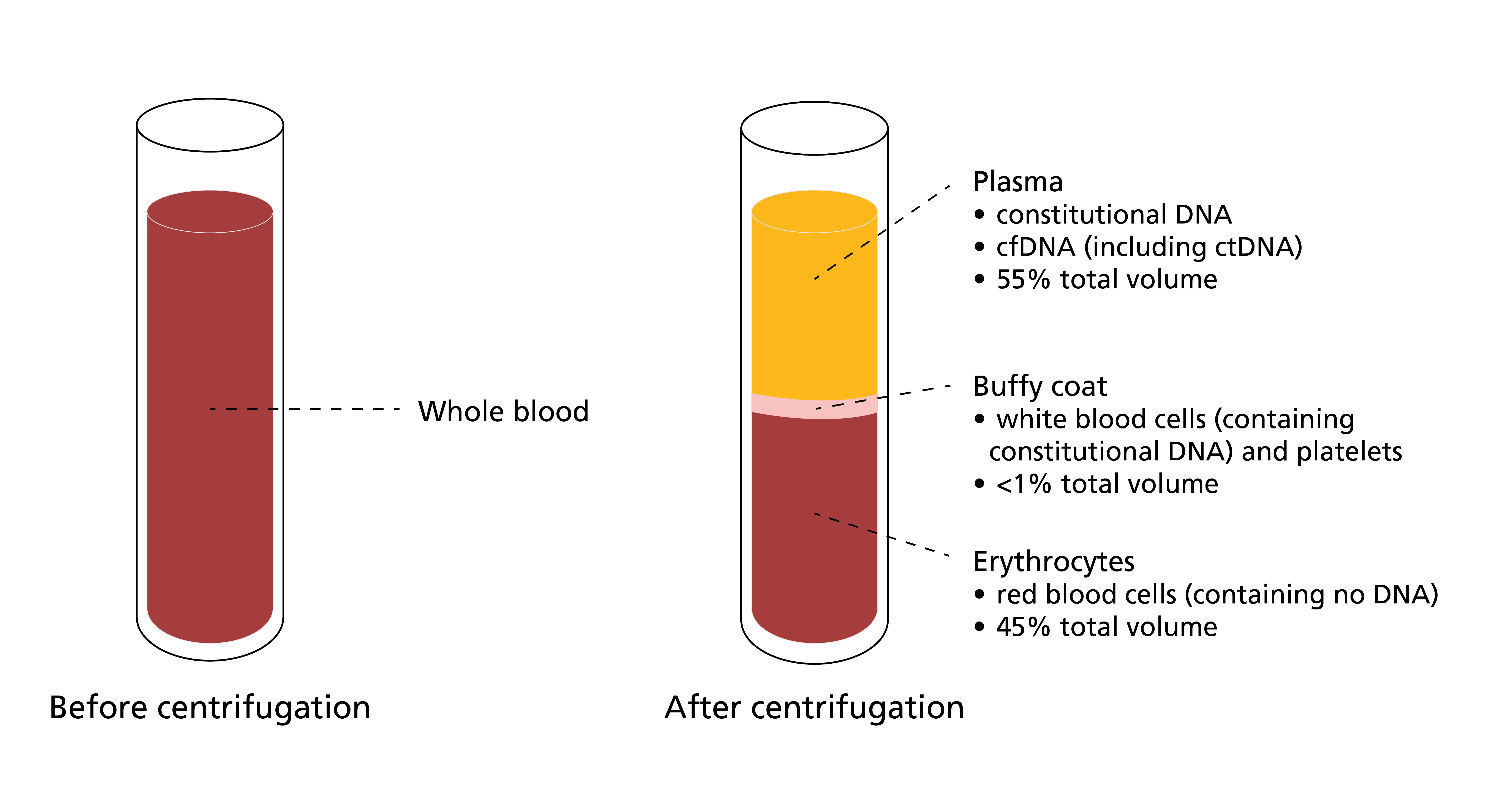
Figure 1: A blood sample before and after centrifugation
Centrifuging a blood sample separates it into three constituent parts: plasma, buffy coat and erythrocytes. Plasma contains both constitutional (germline) and somatic (tumour) DNA, while the buffy coat contains DNA derived from lymphocytes (including variants of constitutional origin and variants reflecting clonal haematopoiesis). The samples can be compared and analysed to determine which variants identified in the DNA extracted from plasma are truly tumour-derived.
Limitations of ctDNA liquid biopsy
There are a number of limitations that need to be overcome before the mainstream implementation of ctDNA analysis in cancer care.
For example, not all tumours shed ctDNA into the blood, even in late-stage disease, which limits widespread application. Results require careful interpretation, with the possibility of false positive results from constitutional (germline) variants or variants arising from hemopoietic cells (clonal haematopoiesis of indeterminate potential).
Applications of ctDNA liquid biopsy
Liquid biopsy has been proposed for a range of applications in clinical practice, including:
- early cancer detection and screening;
- molecular profiling and targeted treatment selection;
- monitoring response to systemic therapy;
- detection of minimal residual disease and monitoring for disease relapse;
- detection of treatment resistance and clonal evolution; and
- prognostication.
Quantification of ctDNA has shown that trends reflect treatment response. Clinical response is associated with reducing levels of ctDNA detectable in the blood.
A rise in ctDNA seen at disease progression has been demonstrated prior to radiological or clinical evidence of relapse (see figure 2). This ‘lag time’ potentially offers a window of opportunity for early intervention and salvage treatment.
Minimal residual disease refers to microscopic cancer remaining following completion of curative treatment, which cannot be detected clinically but carries the potential for disease relapse in the future. Detection of ctDNA after completion of curative treatment has been found to be associated with worse long-term outcomes.
In metastatic solid tumours, the duration of response to targeted therapies is limited by the inevitable development of resistance, and repeat ctDNA monitoring during treatment may be able to detect the emergence of these variants responsible for drug resistance. Studies have shown a correlation between high ctDNA levels and the worst disease prognoses in a variety of different settings and stages.
The use of ctDNA for early cancer diagnosis through screening offers huge potential for patient outcomes, through detection at a stage when the disease is more likely to be amenable to curative treatment. Multi-cancer early detection tests use methylation changes in ctDNA for early detection, including determining the likely cancer site of origin.
ctDNA has also been proposed for tumour site identification in cancers of unknown primary.
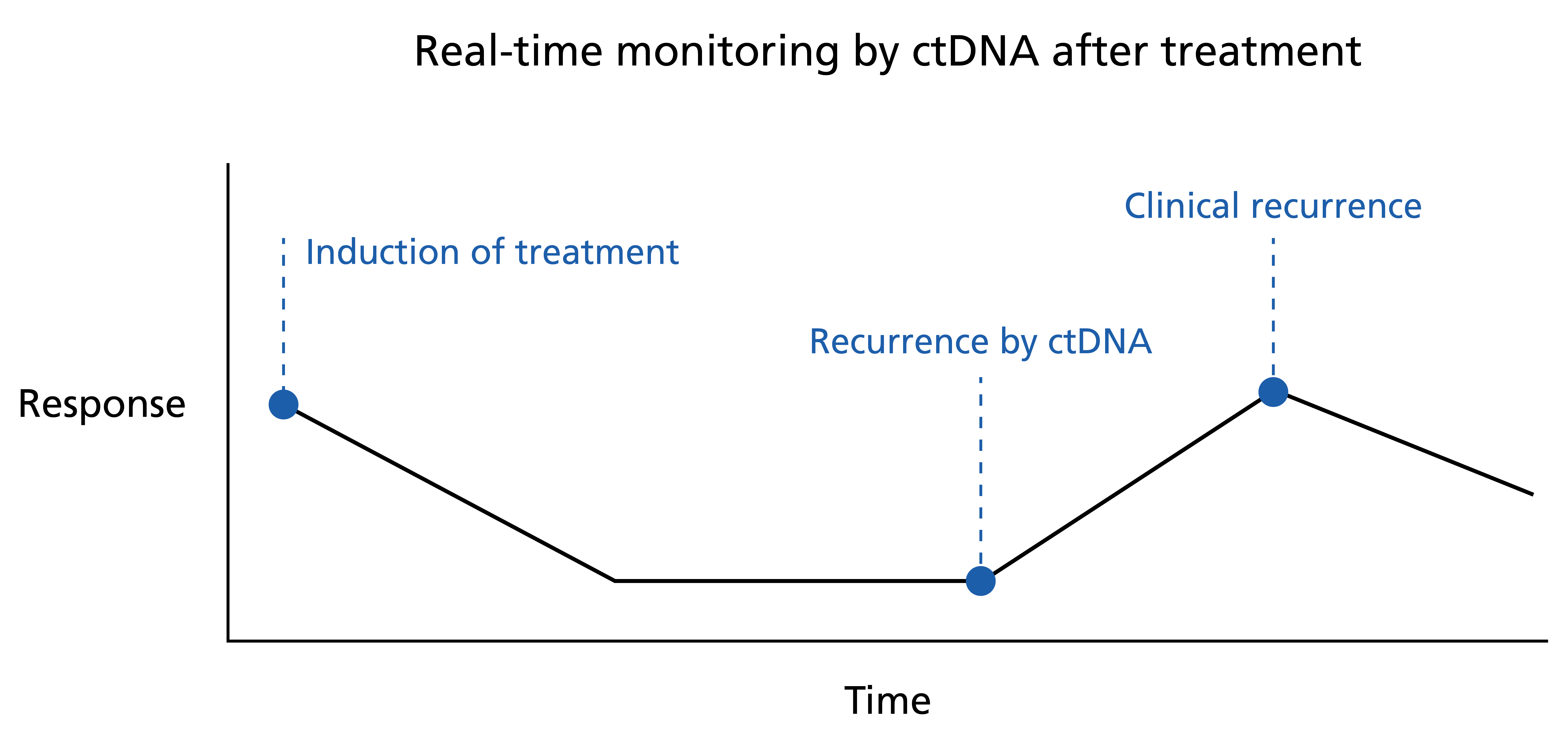
Figure 2: Real-time monitoring by ctDNA after treatment
A rise in ctDNA may be seen before clinical evidence of recurrence, presenting an opportunity for early intervention.
ctDNA analysis
Sequencing of ctDNA can provide genomic information about the underlying tumour; however, there may be discordance between molecular analysis of ctDNA compared with that of the tumour tissue.
Currently, ctDNA sequencing is not widely used for molecular profiling in standard clinical practice, and availability within the NHS is limited. Liquid biopsy requires further clinical validation before being incorporated into routine care, and is generally confined to clinical trials.
ctDNA analysis methodologies
Characteristics of the assay need careful consideration. Analysis methods should be selected depending on the tumour primary, disease extent and clinical application of testing.
Extent of testing
- Sequencing methodologies are broadly classified into polymerase chain reaction (PCR) analysis and massively parallel sequencing (sometimes called next-generation sequencing)-based assays.
- PCR analysis looks for a single variant or a small panel of variants, whereas massively parallel sequencing can incorporate much larger panels of genes.
- Massively parallel sequencing can be used to perform whole genome sequencing, whole exome sequencing or, more commonly, gene panel sequencing. Where panels are used, consideration needs to be given to panel size and the genes incorporated. There are two main approaches to this: tumour-informed and tumour-agnostic.
Tumour-informed and tumour-agnostic testing
- A tumour-informed approach is based on using genomic information obtained by prior sequencing of a tumour sample from the same patient. Benefits include reduced testing costs and faster turnaround times.
- A tumour-agnostic approach applies the same sequencing panel to all patient samples without the requirement of previous tissue sequencing and regardless of the genomic features of the tumour.
-
- Benefits include improved sensitivity through the employment of more sensitive PCR-based methods for plasma sequencing, which may enhance ctDNA detection at low variant allele frequency (VAF).
-
- The need for individualised assay development, however, is more time-consuming and may be more logistically difficult to incorporate into routine care. Moreover, this approach may fail to identify suitable variants owing to issues with tumour sequencing, such as limited tumour cellularity.
-
- Gene panel sequencing for tissue analysis would also need to be considered.
Applications of ctDNA analysis
- ctDNA analysis is able to detect single nucleotide variants and small insertions or deletions, and may be able to detect copy number variants and certain rearrangements.
- In addition to somatic (tumour) variants, ctDNA can be analysed for epigenetic variation, such as methylation changes that control gene expression.
- RNA analysis can be used to investigate the transcriptome.
- Fragment size distribution patterns can also provide prognostic information.
Challenges in analysis of ctDNA
- Analysis can often be limited by total tumour-derived DNA making up a small fraction of the total cell-free DNA proportion.
- VAF refers to the proportion of DNA fragments analysed that display the variant (VAF%). If this proportion is below the limit of detection of the assay, a false negative may result.
- Not all cancers release ctDNA into the circulation. Those that don’t are described as ‘non-shedders’.
- Higher ctDNA levels are usually seen with increased disease burden and at later disease stages.
- Patients with metastatic disease exhibit greater release of ctDNA into the blood, depending on the metastatic distribution of the disease.
- Primary and metastatic spread to the central nervous system is associated with lower rate of ctDNA detection due to the blood-brain barrier.
False positive results
- The term ‘age-related clonal haematopoiesis’ refers to somatic (tumour) variants arising within haematological cell lines. It is seen with increasing frequency with age.
- Identification of variants in genes commonly implicated in haematological neoplasia in an individual without a known diagnosis is referred to as ‘clonal haematopoiesis of indeterminate potential (CHIP)’. Such genes include DNMT3A and JAK2, as well as TP53 and KRAS, in which somatic (tumour) variants commonly arise in solid organ cancers. This provides a source of potential false positives.
- CHIP can be confirmed (and subtracted) by sequencing a matched sample of leucocyte-derived DNA (for example, from the buffy coat) as a control (see figure 1).
- False positive results can also occur owing to the presence of constitutional (germline) variants in ctDNA, and the finding of variants in common cancer susceptibility genes should be followed by appropriate constitutional testing if clinical suspicion of germline origin (based on VAF and the clinical picture) is high. All patients should be counselled on the potential of discovering heritable genetic variants prior to testing.
Current NHS-approved indications for ctDNA
The National Genomic Test Directory lists ctDNA EGFR molecular profiling for hotspot variants in locally advanced or metastatic non-small cell lung cancer to determine eligibility for EGFR inhibitor therapy when tissue testing is not available. Identification of a recognised sensitising variant would make the patient eligible for EGFR-targeted therapy under NHS funding.
Six commercial tests currently have NICE approval:
- EGFR 29 (AmoyDx);
- SuperARMS EGFR T790M mutation kit (AmoyDx);
- Cobas EGFR mutation test v2 (Roche);
- Therascreen EGFR plasma RGQ PCR kit (Qiagen);
- PANAMutyper R EGFR (Panagene); and
- Droplet Digital PCR Dx system (Bio-Rad).
Key clinical trials
Galleri trial
Multi-cancer early-detection tests have been developed for the purpose of cancer screening. They aim to detect the presence of cancer and predict the likely tumour of origin. The GRAIL test screens for more than 50 cancer types by methylation analysis of ctDNA. The Galleri trial is a randomised controlled trial currently underway within the NHS to evaluate the utility of the GRAIL test, alongside existing NHS cancer screening programmes.
PlasmaMATCH
PlasmaMATCH (UK plasma-based molecular profiling of advanced breast cancer to inform therapeutic choices) is a multi-centre, multi-cohort phase-two clinical trial looking at ctDNA for treatment selection in advanced breast cancer. Patients with advanced breast cancer following disease progression were matched to targeted treatments based on variants detected in ctDNA and will be assessed for disease response rate.
TARGET National
TARGET National (tumour characterisation to guide experimental targeted therapy) is a multi-centre trial that performs molecular profiling of ctDNA from patients with advanced solid tumours who have been referred to experimental cancer medicine centres. Results of genomic profiling will be discussed at molecular tumour board meetings with the aim of recruiting patients into phase-one trials on the basis of genetic alterations detected in ctDNA.
Other types of liquid biopsy
Other markers that can be incorporated into a liquid biopsy include:
- circulating tumour cells;
- circulating tumour RNA;
- tumour educated platelets;
- extracellular vesicles; and
- micro-RNA.
Resources
For clinicians
- GRAIL
- MIMS Learning: Circulating DNA as a biomarker for cancer diagnostics
- NICE: Plasma EGFR mutation tests for adults with locally advanced or metastatic non-small-cell lung cancer (PDF, 26 pages)
- NHS Galleri Trial
References:
- Heitzer E, Haque IS, Roberts CES and others. ‘Current and future perspectives of liquid biopsies in genomics-driven oncology’. Nature Reviews Genetics 2019: volume 20, pages 71–88. DOI: 10.1038/s41576-018-0071-5
- Kato S, Krishnamurthy N, Banks KC and others. ‘Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary‘. Cancer Research 2017: volume 77, issue 16, pages 4,238–4,246. DOI: 10.1158/0008-5472.CAN-17-0628
- Pascual J, Attard G, Bidard F-C and others. ‘ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group’. Annals of Oncology 2022: volume 33, issue 8, pages 750–768. DOI: 10.1016/j.annonc.2022.05.520
- Rolfo C, Mack P, Scagliotti GV and others. ‘Liquid biopsy for advanced NSCLC: A consensus statement from the International Association for the Study of Lung Cancer’. Journal of Thoracic Oncology 2021: volume 16, issue 10, pages 1,647–1,662. DOI: 10.1016/j.jtho.2021.06.017
- Wan JCM, Massie C, Garcia-Corbacho J and others. ‘Liquid biopsies come of age: Towards implementation of circulating tumour DNA’. Nature Reviews Cancer 2017: volume 17, pages 223–238. DOI: 10.1038/nrc.2017.7
For patients
- GRAIL
- Macmillan Cancer Support: Personalised medicine
- NHS Galleri Trial