Common aneuploidy testing (QF-PCR)
Rapid testing for the common aneuploidies in prenatal and postnatal samples, and in the investigation of recurrent miscarriage, is routinely undertaken by a rapid polymerase chain reaction (PCR)-based method.
Clinical applications
Aneuploidy is the presence of an atypical number of chromosomes in a cell. Common aneuploidy testing can detect:
- the three most common trisomies: 13 (Patau syndrome), 18 (Edwards syndrome) and 21 (Down syndrome);
- sex chromosome aneuploidy; for example, 45,X (Turner syndrome); and
- triploidy.
The main sample types tested are prenatal chorionic villus or amniotic fluid samples, and products of conception in cases of recurrent miscarriage. Neonates with ambiguous genitalia may also be tested (for sex chromosome aneuploidy), as may postnatal patients with clinical features strongly suggestive of a common aneuploidy (for example, Down syndrome).
How does it work?
Rapid testing for the common chromosomal aneuploidies is performed using quantitative fluorescent-polymerase chain reaction (QF-PCR).
- QF-PCR exploits regions of the chromosomes of interest (13, 18, 21, X and Y) that contain short tandem repeats (STRs). These repeat regions are not themselves associated with disease; however, the number of repeats is highly variable between people, which means that it can be used to identify separate copies of chromosomes present in a sample.
- Primers (short single-stranded DNA with a particular sequence designed to target a unique part of the genome) are available for each STR marker and are labelled with a fluorescent tag. There are multiple STR markers on each chromosome of interest.
- The STR markers are amplified by quantitative PCR, such that the quantity of PCR product reflects the amount of STR marker DNA in the sample (and therefore the copy number of the chromosome it represents).
- The fluorescently labelled PCR products are separated by size using capillary electrophoresis.
- The relative dosage of each STR marker (and therefore each of the chromosome copies in the sample) can then be determined.
How is this applied in common aneuploidy testing?
Aneuploidy can be detected by the presence of atypical ratios or numbers of fluorescence peaks coming from different alleles (chromosomal copies) of each STR (see figure 1).
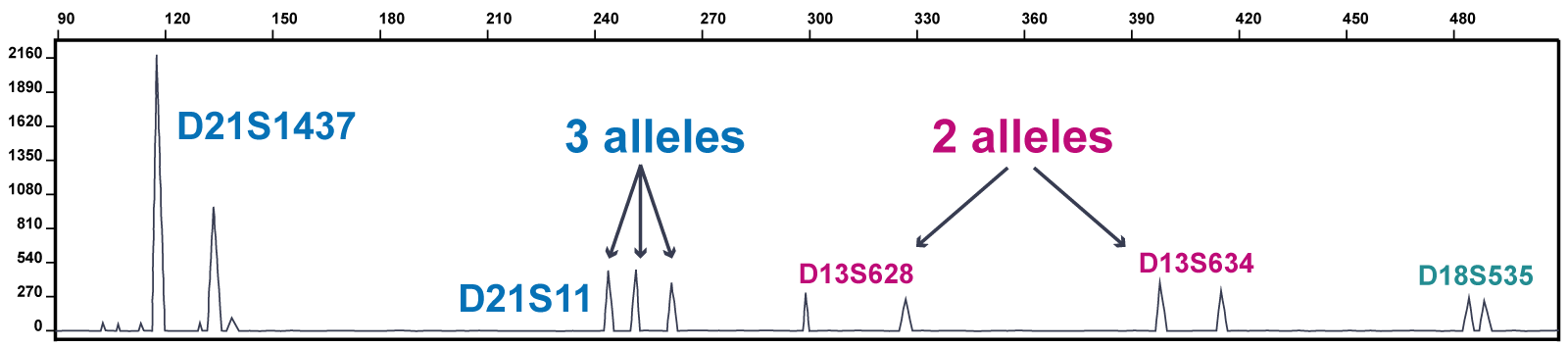
Figure 1: Part of a trace from a QF-PCR test showing trisomy 21
Figure 1 shows part of a trace from a QF-PCR test showing the results from several markers across chromosomes 13, 18 and 21 (the marker labels begin D13, D18 and D21 to indicate the chromosome of origin).
This trace shows trisomy 21. The marker D21S1437 has an atypical peak ratio: the first peak is twice the height of the second peak. The marker D21S11 has an atypical number of peaks (three). The markers from the other chromosomes show two peaks of equal height.
Advantages and limitations of common aneuploidy testing
Advantages
- Common aneuploidy testing is capable of detecting common trisomies, sex chromosome aneuploidy (where appropriate) and triploidy.
- It is rapid: results in as little as 24 hours, fitting within a turnaround time of three days.
- It is cost effective.
- It is robust: it can cope with poor-quality samples and small samples.
- Results do not usually require confirmation by a secondary method.
- In the case of a positive result, the QF-PCR is simply repeated on the same sample for confirmation.
- Karyotyping may be undertaken if there is a technical issue, such as excessive maternal cell contamination.
- Karyotyping may also be undertaken as a follow-up for a detected fetal trisomy, in order to inform recurrence risk by distinguishing that which arises due to non-disjunction of chromosomes (common, low risk) from that which arises due to a Robertsonian translocation (rare, high risk).
- It can detect mosaicism (down to at least 10%).
- It can identify maternal cell contamination (down to at least 10%).
- It can provide information on zygosity in twins.
- If a positive result is obtained, QF-PCR can also provide information on the likely timing of chromosomal non-disjunction. This can help provide an indication of the risk of confined placental mosaicism (when the aneuploidy is present only in the placenta and not the fetus).
Limitations
- Common aneuploidy testing cannot detect any changes that lie outside the target sequence of the STR markers.
- It will not detect balanced rearrangements.
- It may not detect low-level mosaicism.
- In prenatal testing of chorionic villus samples, some types of atypical result are associated with a small but increased risk of confined placental mosaicism, which means that caution must be taken in the interpretation of those samples.
- QF-PCR does not exclude copy number variation other than whole chromosome aneuploidy, nor does it detect single nucleotide variation and, if indicated (for example, fetal anomalies detected on ultrasound), further testing by microarrays or whole exome sequencing may be required.
Practicalities
- The samples required depend on the reason for testing.
- Chorionic villus samples require 12–30 milligrams of cells in transport medium in a sterile, leak-proof container.
- An amniocentesis sample requires 12–20 millilitres of amniotic fluid in a sterile, leak-proof universal container.
- Pregnancy loss or fetal demise requires 2 millilitres of fetal blood in EDTA.
- Products of conception should be in tissue culture media or a dry sterile container. This must not be frozen.
- Neonatal and postnatal babies require a 1–2 millilitre blood sample in a lithium heparin tube.
- Target reporting times from receipt of sample are from 3 to 42 days, depending on the clinical reason for the test.
Resources
For clinicians
- The Association for Clinical Genomic Science: Best practice guidelines for use of quantitative fluorescence-PCR for the detection of aneuploidy, v4 (PDF, 11 pages)
- Online Mendelian Inheritance in Man (OMIM): # 190685
References:
- Mann K and Ogilvie CM. ‘QF-PCR: Application, overview and review of the literature‘. Prenatal Diagnosis 2012: volume 32, issue 4, pages 309–314. DOI: 10.1002/pd.2945