Fluorescent in situ hybridisation (FISH)
Fluorescent in situ hybridisation is a cytogenetic technique used to identify and characterise chromosomal variants.
Overview
Fluorescent in situ hybridisation (FISH) is a technique that uses DNA probes (small molecules of DNA that are designed to hybridise with a particular section of the genome) to detect specific complementary sequences on a chromosome. It is undertaken in situ on chromosomes spread on a slide and visualised by microscopy.
Clinical applications
FISH can provide information on both the copy number of a chromosomal region (whether it is deleted or duplicated) and the chromosomal location of a particular region (whether there is any structural rearrangement, such as an inversion or translocation).
FISH is a labour-intensive and costly technique, and in many areas has been superseded by newer techniques (for example, prenatal aneuploidy screening is now done using quantitative fluorescence polymerase chain reaction (QF-PCR); microdeletions are now tested for by arrays).
Nevertheless, FISH still has a role to play in several areas, outlined below.
- Providing positional information for microarray findings, where this is likely to provide information on the recurrence risk.
- Investigating possible structural or mosaic chromosomal variants (National Genomic Test Directory R298), in particular balanced translocations (which are not picked up by microarray), and clarifying the nature of unbalanced translocations.
- Follow-up testing where ambiguous results are obtained using karyotyping, arrays or common aneuploidy testing; for example, investigation of a marker chromosome (a small fragment of chromosome about which it is unclear which chromosome it is derived from) or investigation of suspected mosaicism.
- In haemato-oncology: identifying the presence of somatic structural variants (for example, fusion genes) that may be drivers of cancer. This may aid diagnosis, estimate prognosis and guide targeted therapy.
How does it work?
A general FISH protocol is described below.
- A fluorescently labelled FISH probe is used that is complementary to the DNA of interest.
- The patient’s cells (which can be, for example, blood, tissue or tumour) are grown in the laboratory (cultured) and arrested in metaphase, and the chromosomes are fixed onto the surface of a glass slide (metaphase FISH). Alternatively, interphase FISH can be performed where cells cannot be cultured, for example on formalin-fixed, paraffin embedded (FFPE) tumour material on a histology slide.
- FISH probes are applied to the slide.
- The FISH probes and the slide are heat treated together to denature both the FISH probe DNA and the genomic DNA into single strands.
- The temperature is lowered to allow hybridisation of the probes to the target region of the chromosome.
- Washes are performed to remove any unbound probe, or any probe bound loosely to non-specific regions.
- The slide is visualised under a fluorescent microscope and the labelled probe fluoresces.
- Multiple probes can be simultaneously hybridised and visualised by using probes with differently coloured fluorescent labels (Figure 1).
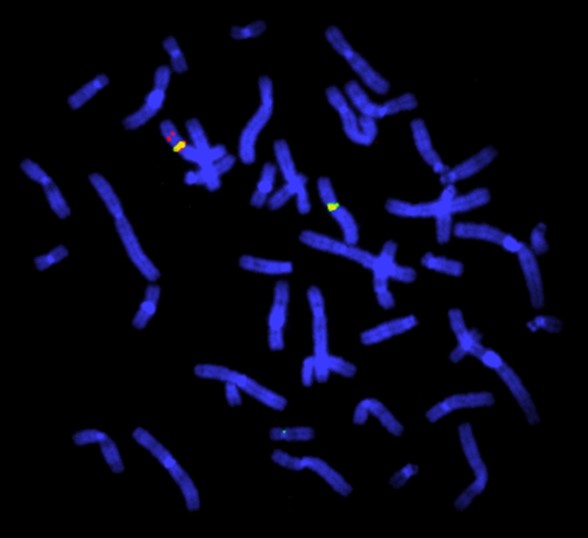
Figure 1: FISH study for deletion in the Duchenne muscular dystrophy gene. This FISH study is of a female heterozygous for a deletion in the Duchenne muscular dystrophy (DMD) gene. The DMD gene probe is red, the X chromosome centromeres are green. There is one X chromosome with no red signal. The chromosomes are blue due to the use of the 4′,6-diamidino-2-phenylindole (DAPI) stain that binds to AT-rich regions. Image by Dr John Crolla: Wellcome Collection, Wessex Regional Genetics Centre. Licensed under 4.0 International (CC BY 4.0)
Advantages and limitations of FISH
Advantages
- It aids interpretation of results from karyotyping and arrays.
- Metaphase FISH can provide positional information (this needs cell culture).
- Interphase FISH can be performed on FFPE slides from tumour material (it provides no positional information, but it can provide information on fusions, amplification and deletions without the need for cell culture).
- It detects whole genome ploidy changes.
- It detects mosaicism.
- It has a fast turnaround time (providing the ability to perform shorter hybridisation in urgent cases).
- Probes are available for almost any genomic region.
Limitations
- It is a targeted test, therefore a limited number of probes can be used at one time with only two or three colours possible.
- Metaphase FISH, which can provide positional information, requires cell culturing so that cells can be synchronised at metaphase.
- Interphase FISH provides no positional information (except for the detection of gene fusions in cancer genetics, for which specific probes are used).
- It is labour-intensive.
- Experienced analysts are needed to interpret FISH images.
- Microduplications may be detected with less sensitivity than microdeletions.
- It cannot detect uniparental disomy.
- Atypical rearrangements may be missed, for example some t(15;17) fusions in hemato-oncology.
- Co-localisation can occur (signals from two FISH probes overlap because the chromosomes happen to be in the same place on the slide and appear as one).
- Cross-hybridisation can occur (the probe binds to regions with repetitive sequences).
Practicalities
- Blood samples must be provided in lithium heparin tubes (typically green-topped). This is different to most genomic tests carried out on blood samples.
- For haematological malignancies, bone marrow aspirate is required. This can be sent in transport media, lithium heparin, or ETDA tubes (typically purple-topped).
- FFPE slides require tumour material (interphase FISH only).
- Target reporting time from receipt of sample is from 3 days to 42 days, depending on the clinical reason for the test.
- It is 3 days for very urgent FISH, for example testing for the PML/RARA (t15;17) in haematological malignancies, acute lymphoblastic leukaemia FISH and rapid aneuploidy testing.
- It is 7 days for urgent testing, for example when testing for mantel cell lymphoma.
Key messages
- FISH detects specific sequences on a chromosome using probes that are visualised using a microscope.
- It can provide information on the copy number and position of a specific sequence, and can also detect amplification of the region.
- It is labour-intensive, but it can provide information that other tests do not with a fast turnaround time possible.
Resources
For clinicians
- Scitable by Nature Education: Fluorescence in situ hybridization (FISH)
- National Human Genome Research Institute: Fluorescence in situ hybridization (FISH)