Short-read sequencing
Short-read sequencing is currently the most commonly used form of next-generation sequencing (NGS) and has a wide range of diagnostic applications. In these types of sequencing, the genome is broken into small fragments (usually 50 to 300 bases) before being sequenced.
Clinical applications
Short-read sequencing is in widespread clinical use. Its clinical applications include whole genome sequencing (WGS), whole exome sequencing (WES), gene panel testing and, increasingly, single-gene testing. Short-read sequencing is also used for many research studies, including the 100,000 Genomes Project.
How does it work?
There are several different short-read sequencing platforms. The basic process of short read sequencing can be found below in figure 1.
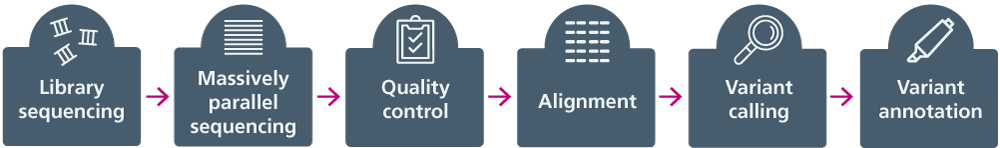
Figure 1: The basic process of short-read sequencing
- Library preparation: DNA is fragmented. The fragments of interest may be enriched by PCR or hybridisation probe capture, and small artificial DNA sequences (adapters) are added, which enable downstream processing and sample identification.
- Massively parallel sequencing: The sequence of bases in many different fragments of DNA is read simultaneously, generating reads usually between 50 and 300 bases long.
- Quality control: The read data are checked to make sure they are worth taking forward to analysis.
- Alignment: Using a reference genome sequence, specialist software maps each read to the specific place in the genome it represents.
- Variant calling: Sequence differences between the sample DNA and the reference sequence are found.
- Variant annotation: The likely effect that a DNA variant will have on a protein and/or a phenotype is predicted.
NGS platforms
Several different short-read NGS platforms are available. They are outlined below.
- Sequencing by synthesis. Detects light emitted when a fluorescently labelled base is incorporated into a growing DNA strand (known as reversible terminator sequencing). This platform is used by Illumina, the most commonly used platform for clinical applications. View this video on Illumina sequencing.
- Semiconductor sequencing (Ion Torrent). Detects the change in pH generated by hydrogen ions being released when a base is incorporated into a growing DNA strand. View this video on Ion Torrent sequencing.
- Sequencing by hybridisation (SOLiD). Now largely displaced by the above methods.
- 454 pyrosequencing. Now discontinued.
Advantages and limitations of short-read sequencing
Advantages
Short-read sequencing carries some of the same advantages as NGS technologies in general. Compared with long-read sequencing, they include:
- being more established in most diagnostic laboratories and therefore widely available;
- higher accuracy; and
- lower cost.
Limitations
Short-read sequencing carries some of the same disadvantages as NGS technologies in general.
The limitations that are specific to short-read sequencing are mostly concerned with the length of the reads generated. As they are short, it may not be possible to map reads to the specific region of the reference genome they came from. If reads cannot be mapped to the reference genome, they may be discarded, leading to a gap in sequencing data.
- Genes with a pseudogene or genes in repetitive regions can therefore be difficult to sequence.
- Repeat expansion disorders can be difficult to detect accurately, especially if the length of the short tandem repeat region exceeds the read length. This is why these are usually tested for separately.
- In addition, large copy number variants and structural variants are not usually detected by short-read sequencing as effectively as they are detected by gold-standard methods such as arrays or karyotyping.
- Methylation changes are not detected, which require methylation studies.
Long-read sequencing is gaining ground in clinical applications as it overcomes some of these issues.
Practicalities
The samples required for short-read sequencing depend on the reason for testing.
- Blood samples are the most commonly used and should be taken into EDTA tubes. EDTA prevents the blood from clotting and inhibits some enzymes that degrade DNA. The volume of blood required varies: from adults, 5–10ml of blood is required; from children 2–5ml is required; and from babies 1–2ml is required.
- Other samples include (check exact requirements with your local laboratory):
- skin and muscle biopsies;
- products of conception;
- placenta samples;
- placental biopsies;
- chorionic villus samples; and
- an amniocentesis sample.
- Saliva samples can be used in some situations, and should be taken into oragene tubes.
- For solid cancers, a biopsy of the tumour is required. The highest-quality DNA and RNA comes from fresh tissue. Formalin-fixed paraffin embedded tissue can be used to obtain DNA and RNA, but the quality is not as high as that obtained from fresh tissue, so some applications may be limited.
- Clinical microbiology sample testing requires appropriate aseptic vials or containers. For specific advice, contact your local clinical microbiology department.
Target reporting time will depend on the reason for testing.
Resources
For clinicians
- Illumina: Introduction to sequencing by synthesis technology
- ThermoFisher Scientific: Ion torrent next-generation sequencing technology
References:
- Goodwin S, McPherson J and McCombie W. ‘Coming of age: ten years of next-generation sequencing technologies‘. National Reviews Genetics 2016: volume 17, pages 333–351. DOI: 10.1038/nrg.2016.49
- Rothberg JM, Hinz W, Rearick TM and others. ‘An integrated semiconductor device enabling non-optical genome sequencing‘. Nature 2011: volume 475, pages 348–352. DOI: 10.1038/nature10242