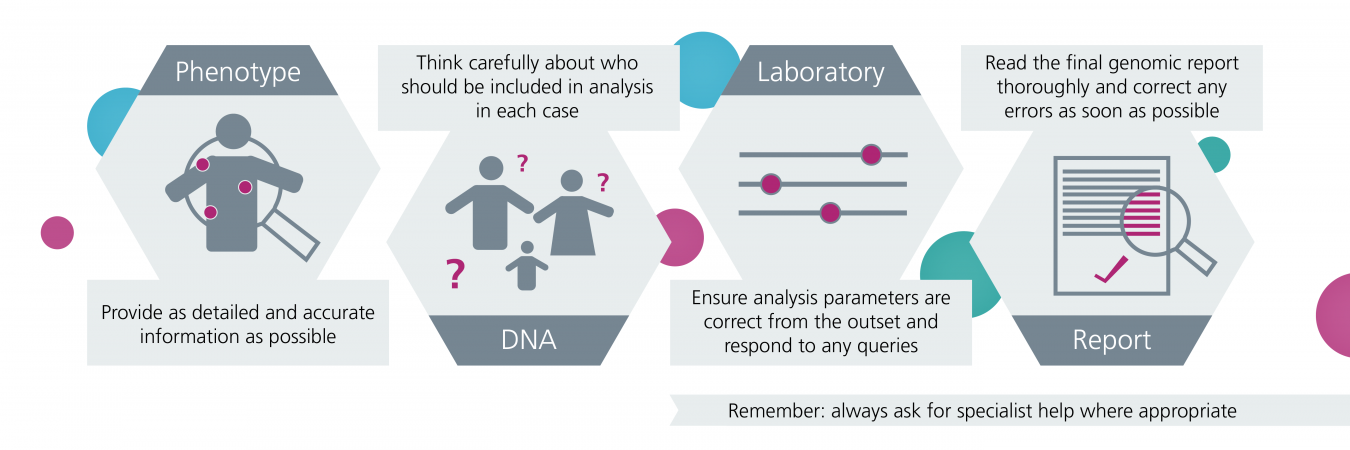
If you’re a clinician requesting genomic testing for a patient, there are some important things for you to know and do in order to maximise the chance that a diagnosis can be found by colleagues analysing the results.
Once a sample has been collected from a patient and genomic sequencing has been performed, scientists are faced with a sea of the individual’s genomic data.
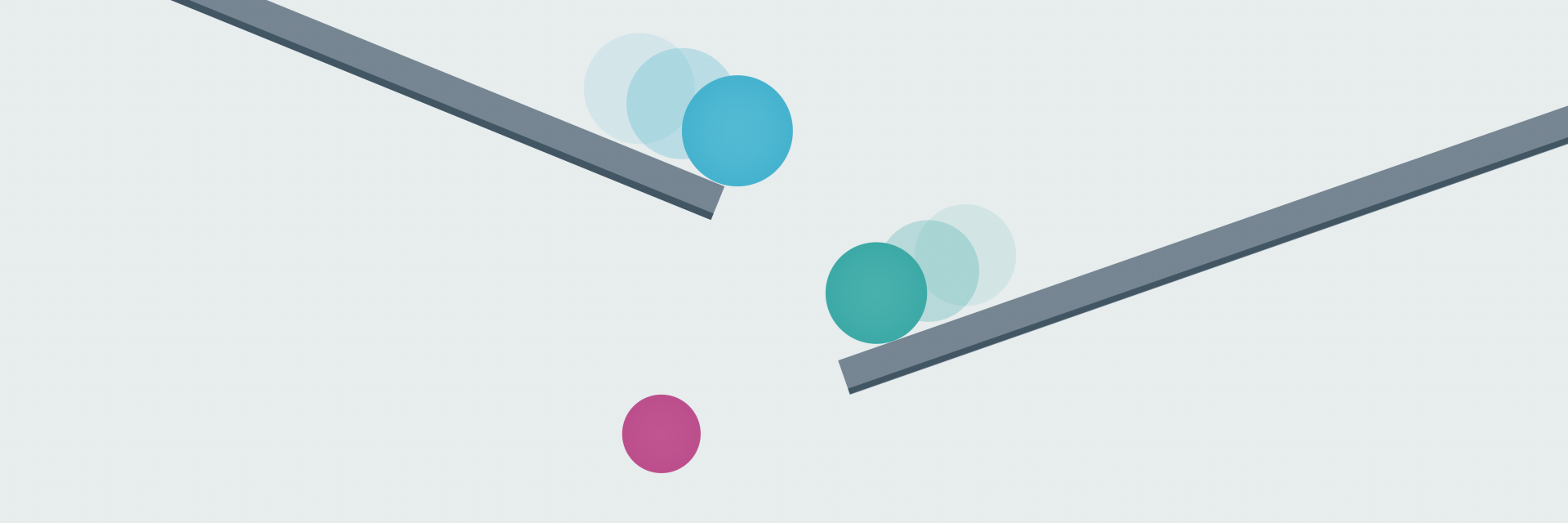
To try and find the genomic variant responsible for disease, they use computer-programming methods and other information to whittle down the genomic data in stages: a process known as variant filtering.
These stages fall into two categories: those that are applied automatically – those that you cannot influence – and those that are based on information you provide: those that you can influence.
Where you come in
Early filtering stages are based on information about the human genome and health at a population level – so-called ‘big data’ – and are out of your control.
For the latter stages, however, the information and samples you provide can make a huge difference to the outcome.
How is the data from genomic sequencing filtered down?
Ordering a test: What you need to know
Summary of key points to consider
Requesting a test: FAQs
1. How do I select patients for genomic testing?
The National Genomic Test Directory for Rare Disease specifies all the available tests and which patients are likely to benefit from a test. In general, the directory focuses on the patient’s context for all tests: If a disorder is likely to have a single genetic cause, and the patient needs a molecular diagnosis to direct their clinical care (or that of their family), then in most cases they will be eligible for a genomic test.
2. What are HPO terms and how do I provide them?
HPO is the Human Phenotype Ontology. It is a standardised way of – a language for, if you like – capturing phenotype information such that it can be used in an automated analysis. There are a number of websites where you can search for the relevant terms to describe your patient’s phenotype, including the one linked above and this one.
Providing HPO terms for unaffected relatives may also be helpful. If the relative’s disease status is listed as unaffected then the analysis will not be limited to variants that relative carries. However, if they have a mild phenotype it may be useful to use this information in the interpretation process.
3. Why do I need to select gene panels for genomic tests?
For some bioinformatics pipelines, particularly where a patient is being tested without their biological parents, it is important to set the appropriate analysis target up front, to reach a manageable number of variants for the final manual review. This is the responsibility of the requesting clinician. NHS panels can be found here, using PanelApp.
If a patient has a complex phenotype, they may need several panels applied to ensure all relevant genes are examined. For example, for a child with developmental delay, regression, microcephaly and seizures it might be appropriate to specify the panels ‘intellectual disability’, ‘genetic epilepsy syndromes’, ‘inborn errors of metabolism’ and ‘mitochondrial disorders’. In general it is more difficult and less efficient to expand the target for analysis after the test has taken place, so providing this information at the pre-test stage is optimal.
4. Should I always try to provide samples and data from other family members?
No. For a child or young person with a complex developmental disorder, providing samples and data from both biological parents increases the chance of making a diagnosis and improves the efficiency of the analysis. For adults with later onset conditions that are often passed down in families, it is better to test only your patient to start with (as long as they have the familial condition).
Providing a broader family history may also be helpful, for example if there is a history suggestive of an X-linked condition.
Additional family samples may be needed after the initial result is available, to check whether a variant of unknown significance is tracking together with the disorder in the family.
5. What is penetrance and how do I know which setting to choose?
Some disorders have ‘complete penetrance’ – that is, a person who carries the disease-causing genomic variant will always develop the condition (sometimes only at a later age: age-dependent penetrance). Other disorders show ‘incomplete penetrance’. This means that some people with the disease-causing variant may never develop symptoms of the condition.
Some analysis pipelines can be set to assume that penetrance is complete or incomplete for a specific analysis. Unless it is very clear that penetrance is highly likely to be complete, it may be best to assume incomplete penetrance, as this covers both possibilities.
6. What do I need to tell my patient about genomic testing?
We have developed a range of resources designed to support health professionals with offering genomic testing to patients.
You may also like to check out our video series ‘Let’s talk about… genomic testing‘.
7. What samples are needed for genomic testing?
Most genomic tests require a blood sample. The sample type is often specified on the test request form. For some specific test types a different sample may be needed. If you need more information, please check the National Genomic Test Directory, or ask the laboratory.
8. Where can I find more resources for facilitating genomic testing?
Check out our web page about the National Genomic Medicine Service, where you’ll find lots of helpful information, online courses and competency frameworks.
9. How do I find my local laboratory?
A full list of the Genomic Laboratory Hubs can be found here.
Many of the hubs also have their own websites where you can find more information, including contact details and how to order a test:
Why might we not have an answer?
Unfortunately, genomic testing may not always provide an answer to explain the clinical situation. Why?